[ English | Japanese ]
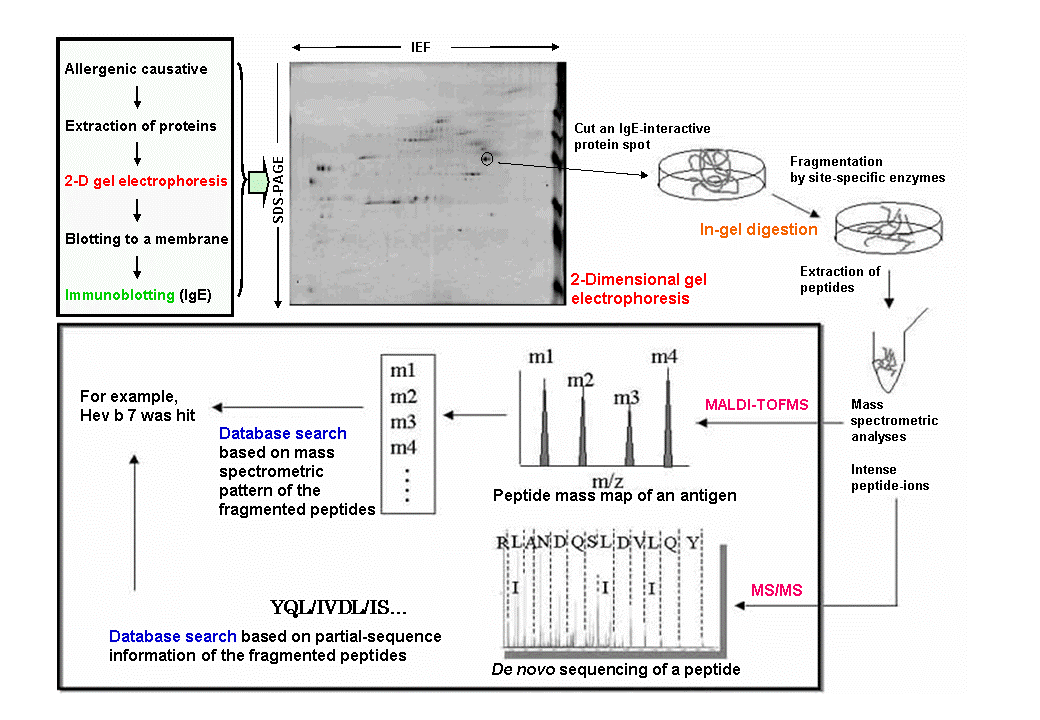 As the sequence information for genes and proteins accumulates in databases, it is becoming easier to comprehensively analyze a series of proteins in question using methods in proteomics like two-dimensional gel electrophoresis and mass spectrometry. Recently, the term "allergenomics" has been proposed for rapid and comprehensive analysis of putative proteinous allergens (allergenome) by applying such a proteomic strategy (Figure). With allergenomics, we can not only detect and assign the putative allergens (proteins specifically interacting with IgE antibodies in a patient's blood) in a short time, but also analyze the quantitative and qualitative change of the antigens, depending on the surroundings and environmental conditions of an allergenic causative [1].
As the sequence information for genes and proteins accumulates in databases, it is becoming easier to comprehensively analyze a series of proteins in question using methods in proteomics like two-dimensional gel electrophoresis and mass spectrometry. Recently, the term "allergenomics" has been proposed for rapid and comprehensive analysis of putative proteinous allergens (allergenome) by applying such a proteomic strategy (Figure). With allergenomics, we can not only detect and assign the putative allergens (proteins specifically interacting with IgE antibodies in a patient's blood) in a short time, but also analyze the quantitative and qualitative change of the antigens, depending on the surroundings and environmental conditions of an allergenic causative [1].Representative allergenomics begins by extracting proteins from an allergenic causative. The extracted proteins are then submitted to two-dimensional gel electrophoresis (Figure). The proteins are first separated with isoelectric focusing (IEF) based on their isoelectric point (pI) using an immobilized pH gradient gel (IPG gel). The focused proteins are then separated again by SDS-PAGE based on their molecular weight. The two-dimensionally spread proteins are then transferred onto a membrane and allowed to react with appropriately pooled serum from patients suffering from an allergy. Thereafter, the antigens specifically interacting with the IgE antibodies are detected. With reference to the results of this immunoblotting, spots of IgE-interactive proteins are picked up from an equivalent two-dimensional gel stained with a dye or fluorescently. The the protein in each gel piece is then site-specifically fragmented by treating it with a proteolytic enzyme like trypsin. This experimental step is called "in-gel digestion". Peptide fragments thus generated are extracted from the gel piece and mass-spectrometrically analyzed (for example, MALDI-TOFMS) to obtain spectra called "peptide mass maps". Moreover, tandem-mass spectra (MS/MS spectra) of several intense peptide-ions on a peptide mass map are recorded. From these mass-spectrometric data, the original IgE-interactive protein, namely an allergen candidate, is assigned by a database search using methods called "peptide mass fingerprint" (PMF) and "peptide sequence tag" (PST).
With current allergenomic methods, comprehensive identification of putative allergens (allergenome) by a standard database search is difficult if the protein or DNA sequences from the corresponding allergenic causative are not fully registered in the databases. However, we can read partial sequences of an IgE-interactive protein from the MS/MS spectra of the fragmented peptides (de novo sequencing). Proteins coming from other species but that are homologous to these partial sequences might be retrieved by database search. Furthermore, we can design oligonucleotide probes with a help of the revealed partial sequences in order to determine the total sequence of the IgE-interactive protein in question through the cloning of its cDNA. It is also important to mention that the actual allergenicity of an identified allergen candidate should be confirmed by other techniques such as a skin prick test or a histamine release test, as the IgE-binding activity of a protein is not necessarily equal to its symptom-eliciting ability.
In the Division of Medical Devices, National Institute of Health Sciences, Japan, we are now studying how allergenomics can be effective for rapid and comprehensive analysis of putative allergens and identifying the difficulties to be solved in the future. Latex allergy and the responsible allergens were chosen as a model case for this purpose [2].
[2] Yagami,T., Haishima, Y., Tsuchiya, T., Tomitaka-Yagami, A., Kano, H. and Matsunaga, K.: Proteomic analysis of putative latex allergens., Int. Arch. Allergy Immunol., 135, 3-11 (2004).